2Department of Thoracic Surgery, School of Medicine, Gazi University, Ankara,
3Department of Thoracic Surgery, Atatürk Chest Diseases and Thoracic Surgery Training and Research Hospital, Ankara
4Department of Medical Biology and Genetics, School of Medicine, Gazi University, Ankara
5Department of Pathology, School of Medicine, Gazi University, Ankara, Turkey DOI : 10.26663/cts.2017.0002
Summary
Background: Non-small cell lung cancer (NSCLC) comprises about 85% of all lung cancers. Although many attempts for early detection and treatment, prognosis of NSCLC is still poor. In recent years the pathways and the genes that play role in lung cancer development were researched widely. PI3K/AKT/MTOR which is thought to be efficacious in the development of many cancer which controls the expression of many genes playing an important role in cell proliferation, metastasis, resistance to apoptosis and angiogenesis. Also the increase in CCND1 expression was shown in several cancer types. The aim of this study is to search the mRNA expression profile of AKT, MTOR and CCND1 genes which has been thought to play role in NSCLC development, and their expression in different pathological stages and histological types of the disease.Materials and Methods: Forty-four NSCLC patients who didn"t get neoadjuvant therapy were included in this study. The samples from tumor and matched normal lung tissue were obtained from resection specimens (lobectomy or pneumonectomy). Total cellular RNA was isolated from the samples. The mRNA expression levels of AKT1, mTOR and CCND1 genes were measured by quantitative real-time polymerase chain reaction (qPCR).
Results: Our findings revealed a statistically significant increase of mTOR expression on mRNA levels (P < 0.05). Although AKT and CCND1 expression slightly increased in malign tissues, these changes in the expression were not significant (P > 0.05). The mRNA expression of mTOR was also upregulated and it was statistically significant for early stage disease and adenocarcinoma subtype (P < 0.05).
Conclusions: Inhibition of mTOR gene expression at mRNA level might be potential target for future treatment strategies of NSCLC.
Introduction
Non-small cell lung cancer (NSCLC) which comprises about 85% of all lung cancers is one of the most common causes of malignancy-related death in developed countries [1,2]. Smoking is the leading risk factor for developing lung cancer, but not all of the smokers develop cancer, suggests that environmental and genetic factors contributes to the risk of cancer development [3]. Although new techniques are developed to detect lung cancer at an early stage, the prognosis is still poor [4]. Hence, development of new therapeutic strategies is still a need for achieving better survival rates.Clinical course of NSCLC can highly differ from patient to patient within same stage and between different age groups. The wide variation in the clinical behavior of similar tumors is related to the difference in the genetic code that underpins the biology of these tumors. Therefore, comprehensive genomic profiling may play a pivotal role in identifying these leading genomic alterations. They could potentially be targeted by molecular agents, which would undeniably change the poor outcome of this devastating disease [5].
Over activation of phosphoinositide 3-kinase (PI3K)/ protein kinase B (AKT), the mechanistic target of rapamycin (mTOR) pathway, plays a pivotal role in tumor growth, invasion, and angiogenesis [6]. AKT is an upstream positive regulatory of the mammalian target of rapamycin (mTOR). In fact, mutation and alteration of gene expressions involved in PI3K/AKT/mTOR pathway are shown to contribute NSCLC development [7].
The key downstream molecular target of PI3K/AKT/mTOR pathway is mTOR [8]. The aberrant expression of mTOR was previously shown in NSCLC [9]. Activation of mTOR results in elevated protein synthesis and G1 cell cycle progression [10]. Furthermore, it was reported that inhibition of mTOR leads to suppression of cyclin D1 and consequently, inhibition of proliferation [11].
The goal of this study was to compare mRNA expression of AKT1, mTOR, and CCND1 (cyclin D1) genes in matched normal and malignant NSCLC tissue samples. Moreover, we also aimed to determine mRNA expression profile of these genes in different pathological stages and histological types of NSCLC.
Methods
Patients CharacteristicsThe study population consisted of 44 newly diagnosed patients with non-small cell lung carcinoma (NSCLC) who underwent curative surgical treatment between March 2011 and December 2011 at Gazi University Thoracic Surgery Department. The study protocol was approved by the Local Ethics Committee of Gazi University Faculty of Medicine and written informed consents were obtained from all patients. The exclusion criteria were, received neoadjuvant treatment (chemotherapy or radiotherapy), radiologic tumor size < 2 cm and tumors which occupy nearly all of the resection material (there is almost no grossly normal tissue). Just after the surgical resection (lobectomy or pneumonectomy) matched tumor and adjacent normal (at least 4 cm apart from the tumor) tissue samples were collected from the resection specimen. The fresh specimens were cut into ∼1 g pieces in operation room and frozen in liquid nitrogen. Then, tissue samples were immediately transferred to -80 °C freezer. All tumor specimens were confirmed by histological examination to contain at least 70% of neoplastic tissue, while all macroscopically normal tissue samples were confirmed to be "normal" by the same pulmonary pathologist.
RNA extraction
Total cellular RNA was extracted using peqGOLD TriFast™ reagent (Peqlab, Erlangen, Germany) according to the supplier"s recommendations. Tissue samples were placed in 1 mL Trizol and homogenized using an Ultra Turrax® T10 homogenizer (IKA Werke GmbH & Co. KG, Staufen, Germany). To remove potential traces of DNA, total RNA samples were incubated with DNase (DNaseI, Roche Diagnostics, GmbH, Mannheim, Germany) for 30 min at 37 °C. Then, all samples were washed with 75% ethanol and resuspended in sterile RNase-free H2O. RNA samples were kept at -80 °C until further analysis. Nanodrop spectrophotometer (NanoDrop ND-1000, Montchanin, DE, USA) was used for measuring RNA concentration precisely.
Complementary DNA (cDNA) Synthesis
Transcriptor First Strand cDNA Synthesis Kit (Roche Diagnostics GmbH, Mannheim, Germany) was used to reverse transcribe cDNA from 1 μg of RNA by the help of random hexamer primer. cDNA reactions were carried out in an Eppendorf Mastercycler EP gradient S thermal cycler (Eppendorf, Hamburg, Germany). Then, all samples were stored at -20 °C until assayed.
Analysis of Gene Expression by mRNA
Gene-specific intron spanning primer pairs and their appropriate probes were chosen relevant to the Universal Probe Library (UPL) Assay Design Center (https://www.universalprobelibrary.com). Primers sequences of the studied genes and UPL probe numbers were listed in Table 1. The crossing points (Cp) were calculated for each transcript using the Light Cycler®480 instrument (Roche Diagnostics GmbH, Mannheim, Germany). Quantitative real-time PCR (qPCR) conditions were as follows; 50 cycles, kept at 95 ºC for 15 seconds and at 60 ºC for 20 seconds and then the samples were cooled to 40 ºC. Gene expression levels of the studied genes were normalized to the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). All qPCR reactions were repeated three times.
Table 1: Primer sequences and UPL probe numbers used in mRNA analysis
Statistical analysis
Relative gene expression results of AKT1, mTOR and CCND1 genes were compared through "REST (2009 V2.0.13)" software using "Pair-wise Fixed Reallocation Randomization" statistical analysis test [12]. The P < 0.05 was considered to be significant.
Results
The details of the patient characteristics are presented in Table 2. The patient group consisted of 41 (93.2%) males and 3 (6.8%) females. The mean age was 61.1 (41-80) years. The exposure to smoking was at an average of 60 pack-years. Only two of the patients had no history of smoking. Histopathologic examination revealed 18 (40.9%) adenocarcinoma and 26 (59.1%) squamous cell carcinoma. According to pathological staging, there were 8 (18.2%) patients with stage 1A, 9 (20.5%) patients with stage 1B, 16 (36.3%) patients with 2A, 4 (9.1%) patients with stage 2B, and 7 (15.9%) patients with stage 3A disease. The types of surgery performed were as follows; 33 (75%) lobectomy, 10 (22.7%) pneumonectomy, and 1 (2.3%) bilobectomy. Mediastinal lymph node dissection was performed in all patients for an effective pathologic staging (averagely 25 lymph nodes were dissected).Table 2: Clinicopathologic characteristics of patients.
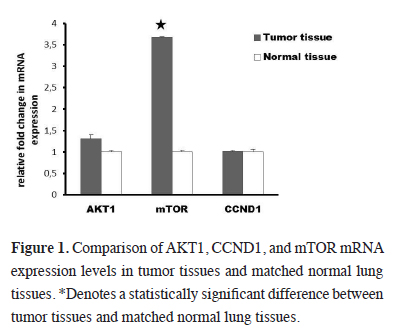
mRNA expression levels of AKT1 in the tumor tissue was 1.1 times higher than in the matched normal lung tissue, but this increase was not significant (P > 0.05). On the other hand the mRNA expression level of mTOR gene was elevated by 2.5 fold (P < 0.05) in tumor tissue when compared to matched normal lung tissue. Also there was a 1.2 fold increase in the expression level of CCND1 in tumor tissue when compared to the matched normal lung tissue, and this increase was not also significant (P > 0.05). mRNA expression levels of studied genes were demonstrated in Figure 1.
 Click Here to Zoom |
Figure 1: Comparison of AKT1, CCND1, and mTOR mRNA expression levels in tumor tissues and matched normal lung tissues. *Denotes a statistically significant difference between tumor tissues and matched normal lung tissues. |
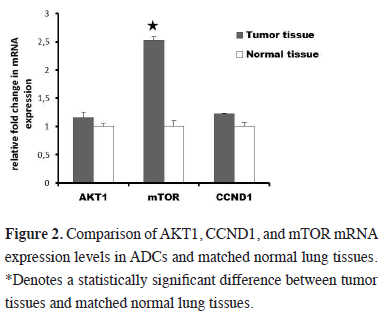
The subtype analyze according to the histologic type of NSCLC revealed statistically significant elevation of mTOR mRNA expression (3.6 fold) in adenocarcinoma (ADC) (P < 0.05) (Figure 2). A slight upregulation of mTOR expression levels is also found in squamous cell carcinoma (SCC) but this increase was not statistically significant (P > 0.05). We failed to find significant differences in AKT1 and CCND1 expression levels among these two histologic types (P > 0.05).
 Click Here to Zoom |
Figure 2: Comparison of AKT1, CCND1, and mTOR mRNA expression levels in ADCs and matched normal lung tissues. *Denotes a statistically significant difference between tumor tissues and matched normal lung tissues. |
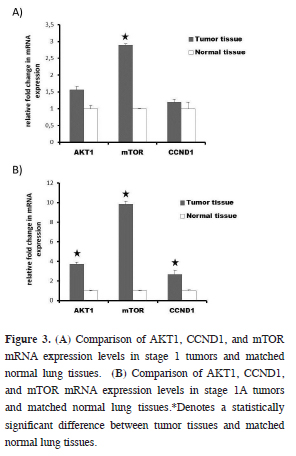
mRNA expressions of studied genes were also analyzed in accordance with pathological stage. mRNA expression levels of mTOR in stage 1 tumors were 2.8 times higher than in the matched normal lung tissue (P < 0.05) (Figure 3A). More detailed analysis revealed that there was a 9.8 fold increase in the expression level of mTOR in stage 1A tumors, when compared to match normal lung tissue (P < 0.05). Two other studied genes, AKT1 and CCND1 were also upregulated 3.7 and 2.6 fold times, respectively (P < 0.05) (Figure 3B). We also found important changes (range 1.9–3.5 fold) for mTOR mRNA expression in other stages between tumor tissue and matched normal lung tissue, but these differences did not make statistical significance (data not shown).
 Click Here to Zoom |
Figure 3: (A) Comparison of AKT1, CCND1, and mTOR mRNA expression levels in stage 1 tumors and matched normal lung tissues. (B) Comparison of AKT1, CCND1, and mTOR mRNA expression levels in stage 1A tumors and matched normal lung tissues.*Denotes a statistically significant difference between tumor tissues and matched normal lung tissues. |
Discussion
NSCLC accounts for 85% of all lung cancers, which adenocarcinomas and squamous cell carcinomas constitute approximately 80% of this type of lung cancer [13]. It is accepted that, the successful discovery, validation and implementation of specific molecular markers for early diagnosis and subsequent timely clinical intervention could vastly improve the appalling mortality rate for this devastating cancer [14].Yue et al. showed an elevated mRNA expression levels of AKT1 gene, as compared to adjacent-tumor tissue [15]. In the present study, the expression of AKT1 mRNA was increased in the tumor tissue as compared to matched normal tissue, but the upregulation was not statistically significant. However, the analysis according to tumor stage showed that mRNA expression level of AKT1 gene elevated by 3.7 fold in stage 1A tumors. No significant differences were found in AKT1 mRNA expression levels among the two histologic subtypes of NSCLC. Up to our knowledge, this is the first study to show the mRNA expression levels of AKT1 in NSCLC in Turkish population.
While there is only one study about AKT1 mRNA expression in NSCLC, several studies were already conducted to analyze mTOR mRNA expression levels in patients with NSCLC. With regard to mTOR expression, it was reported that there was an up-regulation in the expression level of mTOR in tumor tissue as compared to adjacent-tumor tissue [15]. Similarly, Wang et al. showed that mRNA expression levels of mTOR in the tumor tissue were outstandingly higher than in adjacent-tumor tissue [16]. Moreover, Liu et al. reported that mTOR mRNA expression was elevated in tumor tissue as compared to matched normal tissue [9]. These results are similar to our study and show the contribution of mTOR signaling pathway in the development of NSCLC. As far as we know, the current study is also the first to measure mTOR mRNA expression in the tissue samples of NSCLC patients in the Turkish population. We showed an upregulated mTOR mRNA expression levels by 2.5 fold in tumor tissue, when compared to match normal lung tissue.
On the other hand, a previous study has failed to find association between the mRNA expression level of mTOR gene and pathologic type [16]. Similarly, another study showed that the mRNA expression of the mTOR gene tended to increase in patients with advanced stage (stage III) NSCLC, however, this elevation did not reach statistical significance (9). Conversely, we found a relation between upregulated mRNA expression levels of mTOR in stage 1 tumors. We also showed that mRNA expression level of mTOR gene is elevated by 3.6 fold in ADCs.
Detecting of increased mRNA expression levels in stage 1A; a 3,7 fold for AKT1 and a 9,8 fold for mTOR gene, our study suggests that there might be other genes activating mTOR besides AKT1.
Li et al. showed that total CCND1 and its two splice variants (CCND1a and CCND1b) mRNA expression levels were elevated in tumor tissue as compared to nonmalignant tissue at both mRNA and protein levels [17]. In the present study, the level of CCND1 mRNA expression was higher in the tumor tissue than in matched normal lung tissue, but the increase was not statistically significant. Li et al. also demonstrated a relationship between mRNA expression levels of total cyclin D1 and tumor stage of the patients [17]. We found that mRNA expression level of CCND1 gene elevated by 2.6 fold in tumor tissue of stage 1A, when compared to match normal lung tissue. No significant differences were found in CCND1 mRNA expression levels among the two histologic types of NSCLC. According to our knowledge, this is the first study to examine the mRNA expression levels of CCND1 in NSCLC in Turkish population.
Although carcinogenetic effect of smoking habits on lung cancer development is shown via the AKT/mTOR pathway, we are not able to carry out a formal comparison due to the limited number of non-smokers in our study [3]. We believe that further studies with larger number of patients should be conducted to demonstrate such a relation.
Following mTOR-siRNA transfection of NSCLC cells, the suppression of cell proliferation, migration and induction of apoptosis were observed [18]. Hence, RNA interference (RNAi)-mediated transcriptional gene silencing might result in more promising patients" outcome instead of blocking the action of the protein.
The main limitations of our study are the relatively small number of cases and the uneven distribution of female patients as compared to males. But in our country lung cancer incidence is nearly 10 times higher in males and we think our study is truly reflecting Turkish population. Due to freshly resected samples included this study and short patient follow up period; it is not possible to determine the association of mRNA expression of mTOR and prognosis.
As a conclusion detection of aberrant mRNA expression profiles in freshly resected tumor tissue could be helpful to determine real molecular target. Thus, with the selection of patients according to their molecular signature is crucial for achieving the aim of personalized medicine. We can speculate that, inhibition of mTOR gene expression may be used to prevent tumor development because of the presence of higher mTOR mRNA expression at earlier stages of tumor tissues. Short interfering RNA (siRNA)-mediated degradation of mTOR mRNA before translation might take into consideration another therapeutic approach for treatment NSCLC patients. We think this study will be a guide to other researchers who will plan a study on Turkish population. But further studies with larger number of patients should be planned to achieve more definite conclusions.
Declaration of conflicting interests
The author declared no conflicts of interest with respect to the authorship and/or publication of this article.
Disclosure
Partial support for this study was provided by Gazi University Research Foundation (Project code no. 01/2011-21).
Reference
1) Deffebach ME, Humphrey L. Lung Cancer Screening. Surg Clin North Am 2015; 95: 967-78.
2) Hu Q, Zhou Y, Ying K, Ruan W. IGFBP, a novel target of lung cancer? Clin Chim Acta 2017; 466: 172-7.
3) Memmott RM, Dennis PA. The role of the Akt/mTOR pathway in tobacco carcinogen–induced lung tumorigenesis. Clin Cancer Res 2010; 16: 4-10.
4) Luo X, Zang X, Yang L, Huang J, Liang F, Rodriguez-Canales J, et al. Comprehensive Computational Pathological Image Analysis Predicts Lung Cancer Prognosis. J Thorac Oncol 2017; 12: 501-9.
5) Vooder T, Metspalu A. Investigating gene expression profile of non-small cell lung cancer. Cent Eur J Med 2011; 6: 608-15.
6) Bartholomeusz C, Gonzalez-Angulo AM. Targeting the PI3K signaling pathway in cancer therapy. Expert Opin Ther Targets 2012; 16: 121-30.
7) Fumarola C, Bonelli MA, Petronini PG, Alfieri RR. Targeting PI3K/AKT/mTOR pathway in non small cell lung cancer. Biochem Pharmacol 2014; 90:197-207.
8) Caron E, Ghosh S, Matsuoka Y, Ashton-Beaucage D, Therrien M, Lemieux S, et al. A comprehensive map of the mTOR signaling network. Mol Syst Biol 2010; 6: 453.
9) Liu Z, Wang L, Zhang LN, Wang Y, Yue WT, Li Q. Expression and clinical significance of mTOR in surgically resected non-small cell lung cancer tissues: a case control study. Asian Pac J Cancer Prev 2012; 13: 6139-44.
10) Seeliger H, Guba M, Kleespies A, Jauch KW, Bruns CJ. Role of mTOR in solid tumor systems: a therapeutical target against primary tumor growth, metastases, and angiogenesis. Cancer Metastasis Rev 2007; 26: 611-21.
11) Law M, Forrester E, Chytil A, Corsino P, Green G, Davis B, et al. Rapamycin disrupts cyclin/cyclin-dependent kinase/p21/proliferating cell nuclear antigen complexes and cyclin D1 reverses rapamycin action by stabilizing these complexes. Cancer Res 2006; 66: 1070-80.
12) Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 2002; 30: e36.
13) Langer CJ, Besse B, Gualberto A, Brambilla E, Soria JC. The evolving role of histology in the management of advanced non-small-cell lung cancer, J Clin Oncol 2010; 28: 5311-20.
14) Liloglou T, Bediaga NG, Brown BR, Field JK, Davies MP. Epigenetic biomarkers in lung cancer. Cancer Lett 2014; 342: 200-12.
15) Yue W, Wang X, Wang Y. The Relationship between the PI3K/Akt/mTOR Signal Transduction Pathway and Non-small Cell Lung Cancer. Zhongguo Fei Ai Za Zhi 2009; 12: 312-5.
16) Wang L, Xu S, Yue W, Zhao X, Zhang L, Wang Y. Expression and clinical significance of mTOR and PTEN in non-small cell lung cancer. Zhongguo Fei Ai Za Zhi 2010; 13: 717-21.